Interdisciplinary Initiatives Program Round 8 - 2016
Polly Fordyce, Genetics and Bioengineering
Dan Herschlag, Biochemistry
Enzyme catalysis is central to biology: enzymes enhance the speed of all cellular reactions and do so with remarkable specificity. The ability to efficiently engineer novel enzymes would have profound impacts across a variety of fields, from improving industrial processes to designing novel treatments for human health. Current approaches to this engineering involve creating vast libraries of enzyme variants and attempting to select for variants with a desired property (such as converting a substrate of interest to a desired reaction product). However, there is a fundamental limitation to this approach: the number of variants of an enzyme of average length (~200 amino acids) is 20200, a number larger than the number of atoms in the universe. As a result, even the best state-of-the-art selection experiments, which sample 106-109 variants, explore a negligibly small fraction of the possible enzyme sequences, rendering the probability of identifying the optimal variant exceedingly unlikely. Here, we propose an alternate approach: by improving our fundamental knowledge of how enzymes achieve their exquisite catalytic efficiency and specificity, we can use this knowledge to narrow our search to those protein sequences most likely to yield success.
What is needed to gain this fundamental knowledge? Traditional enzymology examines one enzyme at a time, typically mutating individual amino acids in turn to probe their role in catalysis. But each residue does not act alone - rather, residues are interconnected throughout an enzyme, and the ability of residues to function requires these connections to precisely position residues and alter their properties. Therefore, understanding enzyme function will require quantitative examination of thousands of mutant enzymes to reveal the effects of amino acid changes alone and in combination, rather than the handful of variants accessible with traditional biochemical techniques. New technologies are desperately needed.
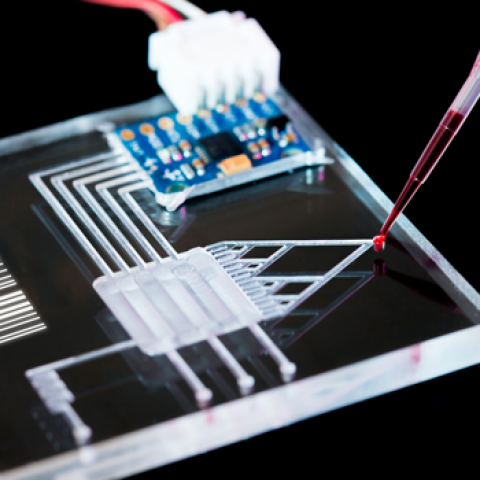
We propose to combine the engineering and microfluidic expertise of the Fordyce lab with the mechanistic and kinetic enzymology expertise of the Herschlag lab to develop a new microfluidic platform capable of a deep kinetic and thermodynamic characterization of thousands of enzyme mutants in parallel in a single experiment. These experiments will allow quantitative examination of how individual mutations affect catalysis alone and in combination, thereby mapping out the full network of residue interactions within an enzyme for the first time and bringing quantitative enzymology into the genomic era. We anticipate that this map will reveal fundamental principles that govern the positioning of active site residues and can be incorporated into current computational techniques for enzyme design. Ultimately, these efforts will improve our ability to generate and manipulate new enzymes for use in technology and medicine.