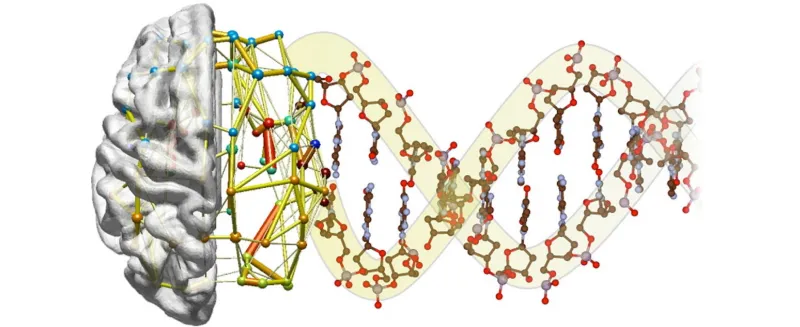
Graphic by Jonas Richiardi: Past neuroimaging studies have defined several “functional networks” in which remote regions of the brain appear to operate in synchrony. A new study from Stanford provides the molecular underpinnings for this theory.
Stanford Medicine News Center - June 11th, 2015 - by Bruce Goldman
A new study by researchers at the Stanford University School of Medicine found that synchronized physiological interactions between remote brain regions have genetic underpinnings.
The research was performed at Stanford but was made possible by collaborations with the Seattle-based Allen Institute for Brain Science and the IMAGEN Consortium, a multicenter European project, said the study’s senior author, Michael Greicius, MD, an associate professor of neurology and neurological sciences.
The study was published June 11 in Science.
An emerging consensus among neuroscientists is that cognitive operations are performed not by individual brain regions working in isolation, but by networks consisting of several discrete brain regions — anatomically connected either directly via white-matter tracts or indirectly through intermediary nodes — that share “functional connectivity,” meaning that activity in these regions is tightly coupled.
Any given functional network is normally most active during the performance of the task associated with that network, as in the case of autobiographical memory (“What did I eat for dinner last night?”). But the synchronous activity of component regions persists when networks are idling. Well over a dozen functional networks have been identified via a technique called resting-state functional magnetic resonance imaging, said Greicius, who is the medical director of the Stanford Center for Memory Disorders.
In resting-state fMRI scans, the individual is asked to simply lie still and relax for several minutes. The results of these scans indicate that even at rest, the brain’s functional networks continue to hum along at their own distinguishable frequencies and phases, like different radio stations playing simultaneously, but quietly, on the same radio.
Digging into fMRI images
However, whether resting-state fMRI-derived images, which measure local blood flows in different places throughout the brain, actually reflect neuronal activity has been controversial.
“There’s been some skepticism regarding the validity of resting-state network activity,” said Greicius. “We wanted to dig deeper and get to the molecular underpinnings of these imaging results, which indicated that the brain maintains its exquisite functional-network architecture even at rest.”
To start, Greicius and his colleagues computationally blended resting-state fMRI data they’d obtained from eight-minute scans of 15 healthy adults whose sole instructions were to lie still and relax. This enabled them to pinpoint numerous well-delineated functional networks.
Hoping to find genes that might promote or at least be involved in functional connectivity, the investigators next sought gene-expression profiles — measurements of activity levels of each of the human genome’s approximately 20,000 known genes — of regions within corresponding functional networks.
There’s no noninvasive way to obtain gene-profile expressions of brain tissue in living humans. But Jonas Richiardi, PhD, a postdoctoral scholar in Greicius’ lab now at the University of Geneva in Switzerland, made use of massive amounts of carefully annotated and meticulously archived data derived from the Allen Institute’s collection of six post-mortem human brain samples. The institute’s scientists have obtained gene-expression profiles of several hundred tissue samples excised from specific locations throughout the brain. Richiardi shares lead authorship of the study with neurology instructor Andre Altmann, PhD, who was also a postdoctoral scholar during the study’s duration.
Greicius and his colleagues narrowed their focus to cortical areas associated with four functional networks that are all well characterized in the imaging literature; consist of discrete, noncontiguous regions in both hemispheres; and are well represented in the Allen Institute’s human-brain database. Along with the default-mode network associated with autobiographical memory, they looked at gene-expression profiles in component regions of the brain’s sensorimotor, visuospatial and salience (emotion) networks.
Zeroing in on gene activity
The researchers were hunting specifically for a set of genes whose expression rose or fell in a more synchronized fashion from region to region within a given network than between networks or outside any network. Using sophisticated statistical methods, they identified a set of 136 genes that showed a correlated pattern of gene expression in regions within each network.
These 136 genes weren’t specific to any single network, Greicius noted. Rather, “any one of these genes that was being expressed at a high, intermediate or low level in one region of any network, regardless of which network you’d picked, was also being expressed at corresponding levels in the other regions of that network,” he said.
Importantly, a number of these genes encode proteins that aid in nerve cells’ signature activity: propagating electrical impulses. Some are ion channels, which maintain and modulate voltage differences across nerve cells’ outer membranes. Others are found at the junctions where one nerve cell in a circuit contacts another.
The Stanford team validated their findings by turning to another database. The IMAGEN Consortium has conducted widespread imaging, cognitive and genetic tests on 14-year-olds in an effort to predict who’s at high risk of encountering problems such as substance abuse by age 16. Among other things, the IMAGEN database contains detailed information on tiny variations from the norm in subjects’ genomic sequences. Altmann spearheaded an analysis of the variants present in the 136 genes of interest in 259 healthy adolescents. These subjects’ network-connectivity strength was determined, in part, by the genetic-variant profiles of these 136 genes.
Additional experiments using tissue samples obtained from two additional data sets, the Allen Institute’s mouse-brain and mouse-connectivity atlases, confirmed and amplified the findings from research on human brains. The reliance on large, shared data sets was another important feature of the study and, Altmann said, “highlights the value of making scientific data freely available. We had an idea and found collaborators willing to share their painstakingly collected data."
The identification of functional-connectivity-associated genes sets the stage for targeted clinical applications, such as finding out how neurodegeneration propagates within a network.
“Our work holds potential implications for a number of neuropsychiatric disorders,” said Richiardi.
Evidence suggests, for instance, that Alzheimer’s disease spreads from one brain region to the next within the brain’s so-called default-mode network, which is activated when a person is recalling recent autobiographical events. Resting-state imaging holds exceptional potential in cases where task-based fMRI isn’t applicable. Alzheimer’s patients, for example, have difficulty focusing on memory-based tasks. Future work will focus on genes whose expression is correlated within one network, but not in other networks. Focusing on default-mode network-specific genes, for example, may lend novel insights into Alzheimer’s disease.
Another Stanford co-author of the study is postdoctoral scholar Anna-Clare Milazzo, PhD.
The experiments reported in the study were funded by the National Institutes of Health (grant R01NS073498), the Allen Institute, the Feldman Family Foundation, the IMAGEN Consortium and a Marie Curie Fellowship from the European Union.
Information about Stanford’s Department of Neurology and Neurological Sciences, which also supported the work, is available at http://neurology.stanford.edu/.

