Photo by Kateryna Kon, Shutterstock.
Stanford Medicine Scope - April 20th, 2017 - by Bruce Goldman
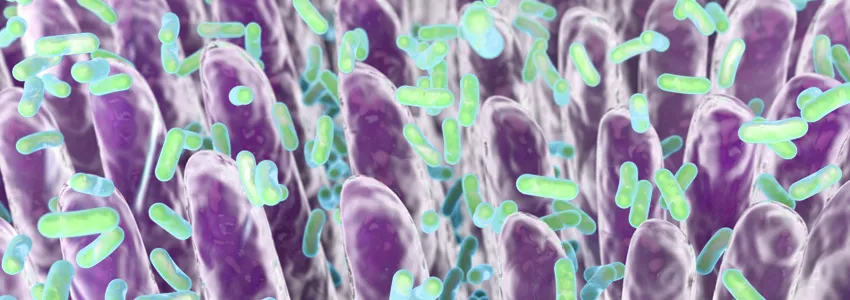
In a mouse study published today in Cell, Stanford microbiologist Justin Sonnenburg, PhD, and his colleagues have been able to simultaneously visualize multiple bacterial strains in lab mice’s gut by making them express unique combinations of fluorescent proteins. This approach lets researchers pinpoint the location of bacteria in the gut based on the rainbow of colors they’re induced to emit.
Gut microbes play wide-ranging roles in health and disease. But techniques for probing the relationship between microbial activity and host physiology have been largely unavailable.
“These new tools open the door to new types of studies to better understand our gut commensal bacteria and to define how they can be engineered for therapeutic purposes,” Sonnenburg said.
Advances in sequencing technology have permitted in-depth characterization of bacterial species found in the gut, but tools for manipulating the hundreds of species of microbes that make their homes in the gut have lagged far behind. Although such techniques have been developed for model organisms used in research, such as E. coli, they don’t work in more relevant microorganisms such as Bacteroides, the most abundant genus in the guts of people in the United States.
Sonnenburg and his team developed a way to engineer Bacteroides, making it possible to simultaneously track multiple bacterial strains in the gut. The researchers inserted — into the bacterial genome — a panel of synthetic DNA sequences that can initiate the activation of particular genes.
Using this panel of DNA sequences, the researchers genetically engineered six different Bacteroides species to produce unique combinations of a red fluorescent protein and a green fluorescent protein. They introduced the engineered species into mice that had been raised in a germ-free environment, and after two weeks, they analyzed sections of colon tissue using a fluorescence microscope to pinpoint the location of the bacteria in different parts of the gut.
In a separate experiment, Sonnenburg and his team genetically engineered two Bacteroides strains to produce either the green or the red fluorescent protein. They then introduced the red-fluorescing strain into the mouse gut, followed by the green-fluorescing strain one week later. The “red” strain successfully colonized the colon and out-competed the johnny-come-lately “green” strain, especially in tube-like compartments of the gut called crypts. The team’s findings demonstrate that colonization of these specific intestinal structures is a key step enabling longer-term gut residents to out-compete invading species.
In future research, Sonnenburg plans to develop these tools to engineer bacteria to produce proteins — including ones with therapeutic properties — at a precise time or location.
However, he cautions: “Before gut commensals can be engineered for therapeutics, it will be important to develop methods of safely and reliably colonizing the human gut, which will require more research.”

