Photo by Vignesh Ramachandran: Zahid Hossain, Ingmar Riedel-Kruse, Nate Cira and Seung Ah Lee created biotic games that allow people to interact with cells. They also created a robotic biology lab capable of carrying out remote-controlled experiments.
Stanford Report - April 23rd, 2015 - by Erin Digitale
In the 1950s, computers were giant machines that filled buildings and served a variety of arcane functions. Today they fit into our pockets or backpacks, and help us work, communicate and play.
"Biotechnology today is very similar to where computing technology used to be," said Ingmar Riedel-Kruse, an assistant professor of bioengineering at Stanford.
"Biological labs are housed in big buildings and the technology is hard to access," he added. "But we are changing that. We are enabling people to interact with biological materials and perform experiments the way they interact with computers today. We call this interactive biotechnology."
Riedel-Kruse and his team have created three related projects that begin to define this new field of interactive biotechnology.
Two of these projects will be unveiled this week at the Computer Human Interactions (CHI) conference in Seoul, South Korea, while the third was recently published in PLoS Biology.
In one project the team created an arcade- style kiosk that allowed visitors to The Tech Museum in San Jose to interact with living cells like fish in an aquarium.
In a second and similar effort, Riedel-Kruse developed a project to teach students how to design bioengineering devices by creating so-called biotic games using cells. This class also touched on the ethical principles of interacting with microorganisms for educational or entertainment purposes.
One of the educational projects in the Riedel-Kruse lab.
In his third and most far-reaching project, Riedel-Kruse created a robotic biology cloud lab capable of carrying out remote-controlled experiments.
"We call these robots Biotic Processing Units or BPUs," said Zahid Hossain, the Stanford doctoral student who worked with Riedel-Kruse on this third project.
"A BPU is an instrument that can hold and repeatedly stimulate biological materials, such as cells, and measure the biological responses," Hossain said. "It is the key enabler of interactive biotechnology."
This cloud lab is being presented at the computer conference in South Korea. The intent was to enable students and scientists to send instructions to a robotic lab and get back experimental results – the way people interact with cloud-based data sites today.
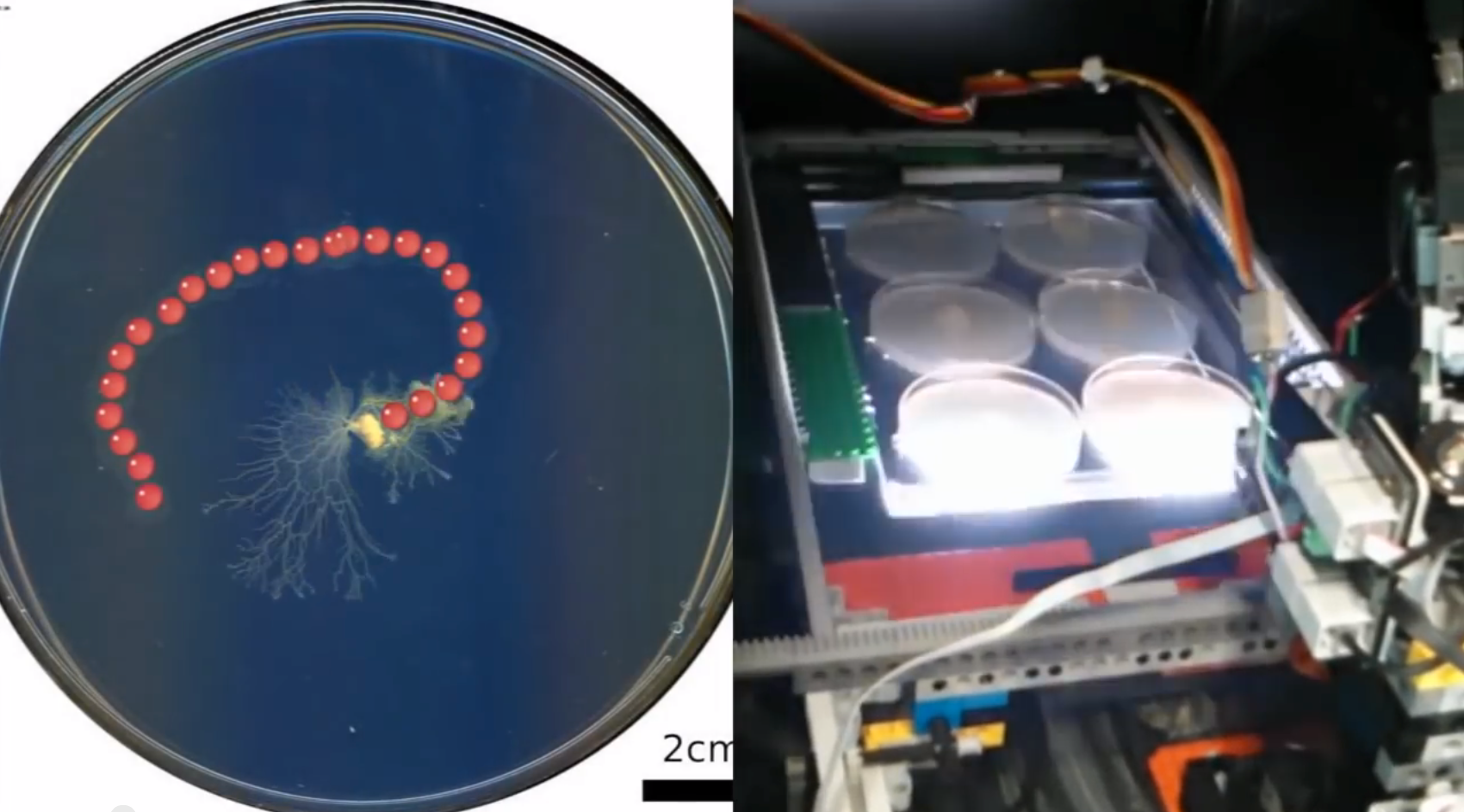
Hossain constructed this prototype BPU by using LEGO Mindstorms to create a liquid-handling robot. This robot traveled over a flatbed photo scanner. The scanner held petri dishes containing the slime mold Physarum, which eats oatmeal.
The researchers incorporated this BPU as a lab component in a graduate level theory class. Using remote-control interfaces on their smartphones, students ordered the robot to drop oatmeal onto specific petri dishes. The software allowed them to choose different droplet patterns. The scanner recorded how the Physarum followed each trail of oatmeal dots by "sniffing out" chemical cues in the petri dishes. Chemotaxis is the scientific term used to describe how microorganisms respond to chemical stimuli in their environments.
For this project Riedel-Kruse's team built three BPUs, each holding six petri dishes. All three units were housed in a server rack typically found in a cloud computer site.
"Our prototype BPUs supported 18 users and allowed us to assess the scalability of cloud labs," Hossain said. "I want to see advanced BPUs supporting many different types of experiments and thousands of different users."
This cloud lab project won an honorable mention as a best paper at the computer conference.
The museum kiosk project, led by Seung Ah Lee, a postdoctoral fellow in the Riedel-Kruse lab, will also be presented in South Korea.
Lee explained that the kiosk allowed museum visitors to interact with Euglena, a freely swimming microorganism that typically lives in ponds.
Like plants, Euglena can convert sunlight into sugar through photosynthesis. The interactive display capitalized on the organism's responses to light. In their interactive kiosk, the Euglena inhabited a micro-aquarium that was essentially a specially configured slide mounted between a video microscope and an image projector. This slide, or micro-aquarium, was another instance of what the researchers call a BPU.
This self-contained micro-aquarium was connected to a touch-screen computer display. Museum visitors could use blue, green or red light to draw patterns on the screen and observe how the Euglena reacted. The microorganisms avoided blue light, so drawing a circle around one of the microbes would trap it, which became the name for one of the scientific mini-games the kiosk offered.
Riedel-Kruse also used the light-sensitive Euglena as the model organisms in the class he taught on biotic game design. Nate Cira, the bioengineering doctoral student who led that effort, said the goal was to create a biotech version of popular robotic and video game challenges. He said the team plans to create low-cost kits that would allow hobbyists to construct their own interactive micro-aquariums.
To assess the educational value of these projects, Riedel-Kruse worked with Stanford education Assistant Professor Paulo Blikstein. The biotic game design class was jointly taught with bioengineering Professor Stephen Quake.
Riedel-Kruse said he thinks that interactive biotechnology is a necessary and inevitable consequence of the maturation of the life sciences that will profoundly affect society, just as computing technology has.
"The obvious next application is online education at scale that includes true biology experiments, also opening new opportunities for learning-research. And cloud labs can change how we work as scientists." Riedel-Kruse said. "Ultimately, I hope these interactive media make everyone more understanding and comfortable about what microbiology and biotechnology really is."



